Media Exposure
-

Marine Virus-Mediated Processes Impact Carbon…
A groundbreaking study from Bar-Ilan University, led by Dr. CHana Kranzler of… -

A €150,000 Grant Offers New Hope for Young Ca…
How Cutting-Edge Medical Research is Paving the Way for Fertility Preservation… -
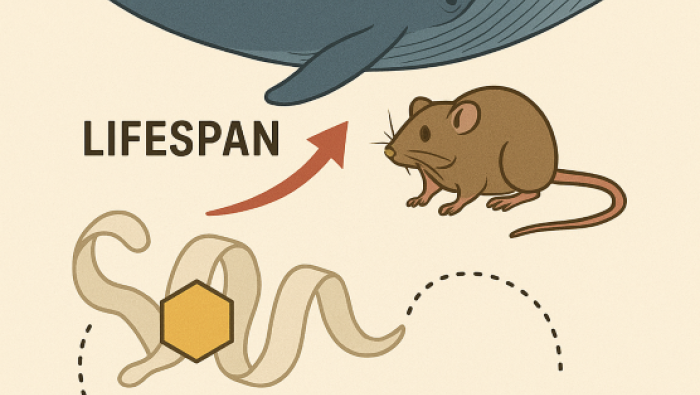
Bar-Ilan University Evolutionary Analysis Rev…
Over recent decades, humanity has witnessed a remarkable and continuous… -
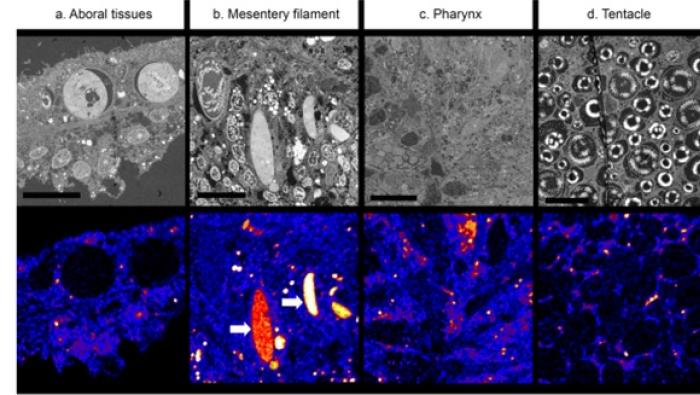
A method to disentangle and quantify host ana…
A wide range of organisms host photosynthesizing symbionts. In these animals… -
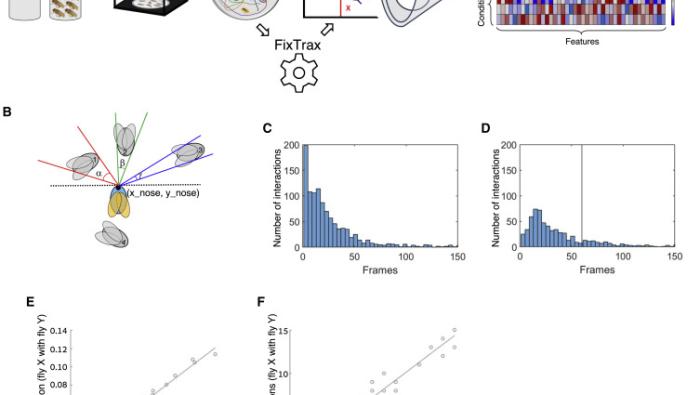
Early Life Experience Shapes Male Behavior an…
Living in a group creates a complex and dynamic environment in which behavior… -
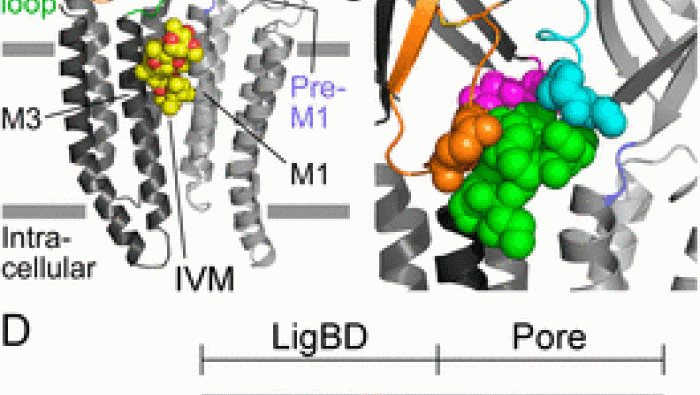
Subunit stoichiometry and arrangement in a he…
-
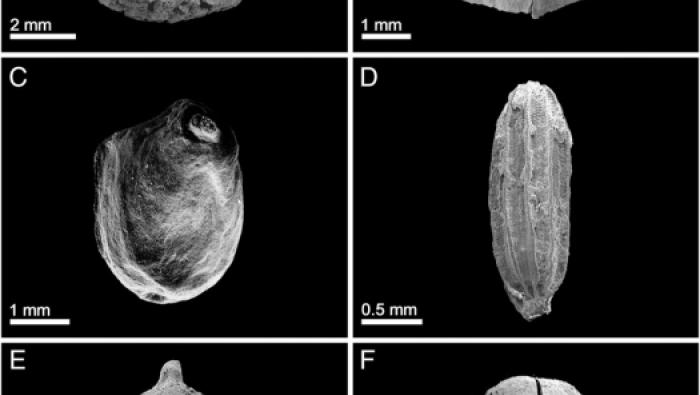
The plant component of an Acheulian diet at G…
-
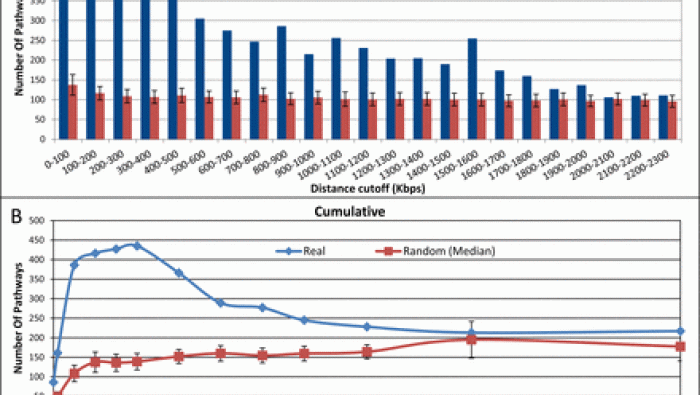
How far from the SNP may the causative genes…
-
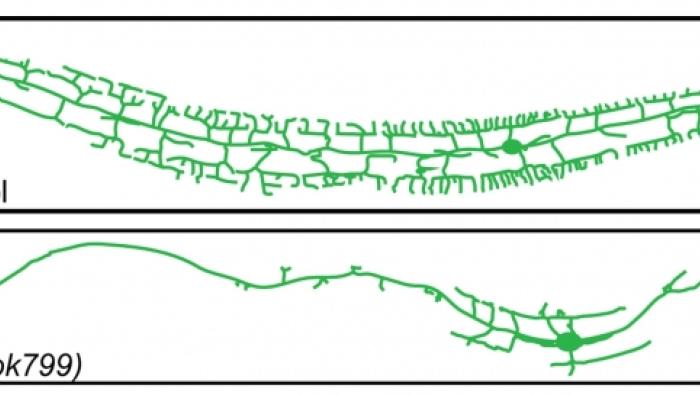
Reduced Insulin/Insulin-Like Growth Factor Re…
-
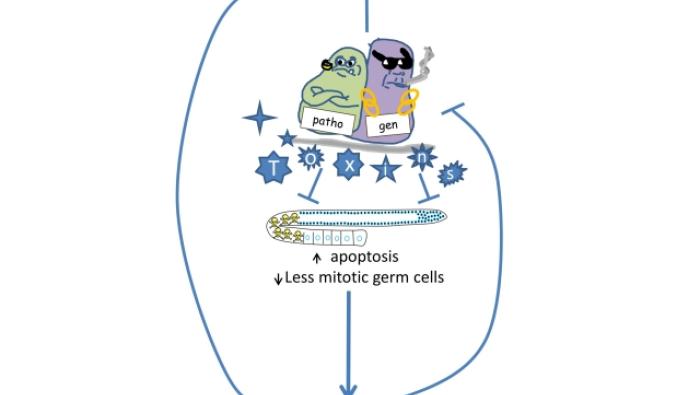
Innate immunity mediated longevity and longev…
-
Dynamics and Transport of Nuclear RNA
-
De-novo protein function prediction using DNA…